Do you have questions about running a computational process for genomics data? Is the process new to you, or are your systems unable to process a job of this scale? We are available to help. Computational genomics consulting is available to all faculty, staff, and students at UTIA. The consulting is offered by Ryan Kuster at rkuster@utk.edu.
Consultation Fees
The first consultation meeting is free of charge and will be used to develop a plan of action catered to your areas of need. The hourly fee for general consultation and analysis is $80/hour (please contact us for external pricing).
Data Analysis Support
Past consulting has included these topics:
- Genome or Transcriptome Assembly
- Gene Annotation
- Differential Expression Analysis
- Variant Calling (SNPs, InDels, SVs)
- Generating SSR Markers
- Small RNA Analysis
- How to Order Sequencing (Platform, Depth, Price, etc.)
*If what you need isn’t on the list, we are still happy to meet with you and talk through possible support options.
Training
- Software Installation on Linux
- Using ISAAC HPC
- R and Python Scripting
- Running containers with Singularity
- Data management and version control with Git/GitHub
- Workflow Development with Nextflow
UTIA Computational Equipment
We are excited to announce the UTIA Computational Resources, which consists of two new computers dedicated to intensive computational projects:
- Centaur (centaur.ag.utk.edu): 96 Cores, 512 Gb RAM, 200 Tb iSCSI storage.
- Sphinx (sphinx.ag.utk.edu): 64 Cores, 512 Gb RAM, 70 Tb SSD storage.
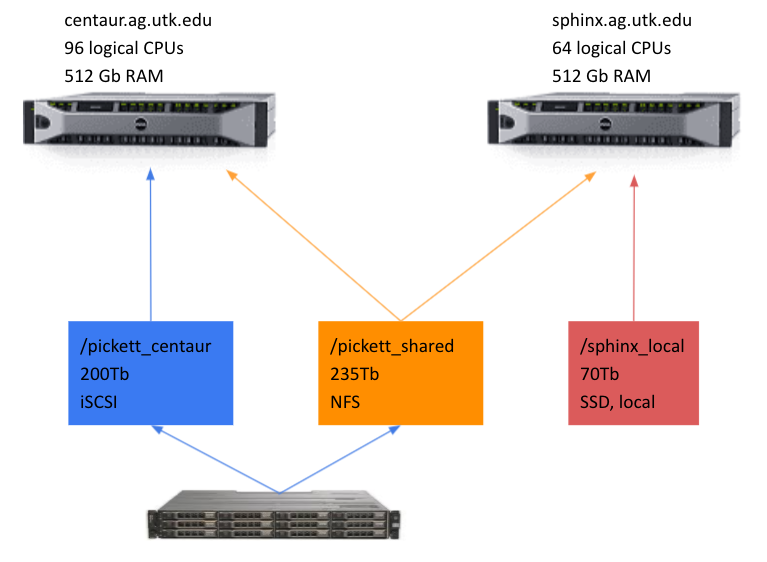
Our goal is to provide a more flexible system than what is currently available with ISAAC; you will not be required to run jobs through a scheduling system.
We require anyone who wants to use these resources to go through an orientation presentation. Contact us at mstaton1@utk.edu to schedule your orientation. General documentation, tutorials, and FAQs can be found on our GitHub wiki.
Resources
- Courses, Workshops, and Journal Clubs
- Check back often for future courses, workshops, and journal club offerings.
- Equipment
- UTIA
- UTIA OMICS Hub
- UTIA Genomics Center for the Advancement of Agriculture
- EPP 622 Bioinformatics Applications – offered Fall of even years
- UTK